Filtering and QC of VCFs
Tools used: dumpSTR, qcSTR
This vignette shows how to use dumpSTR to filter a VCF and qcSTR to visualize some basic QC metrics. For this example, we use the file trio_chr21_popstr.sorted.vcf.gz available at https://github.com/gymrek-lab/TRTools/tree/master/example-files. This file was generated on samples NA12878, NA12891, and NA12892 using popSTR.
First, let’s perform some filtering on the VCF:
dumpSTR --vcf trio_chr21_popstr.sorted.vcf.gz --popstr-require-support 2 --popstr-min-call-DP 10 \
--out popstr-filtered --min-locus-callrate 1
bgzip -f popstr-filtered.vcf
tabix -p vcf popstr-filtered.vcf.gz
This command filters calls with depth of less than 10 or with alleles supported by less than 2 reads, and loci with any missing genotypes. Now, we can run qcSTR on the filtered VCF:
qcSTR --vcf popstr-filtered.vcf.gz --out popstr-qc
This will output the following files:
popstr-qc-sample-callnum.pdf
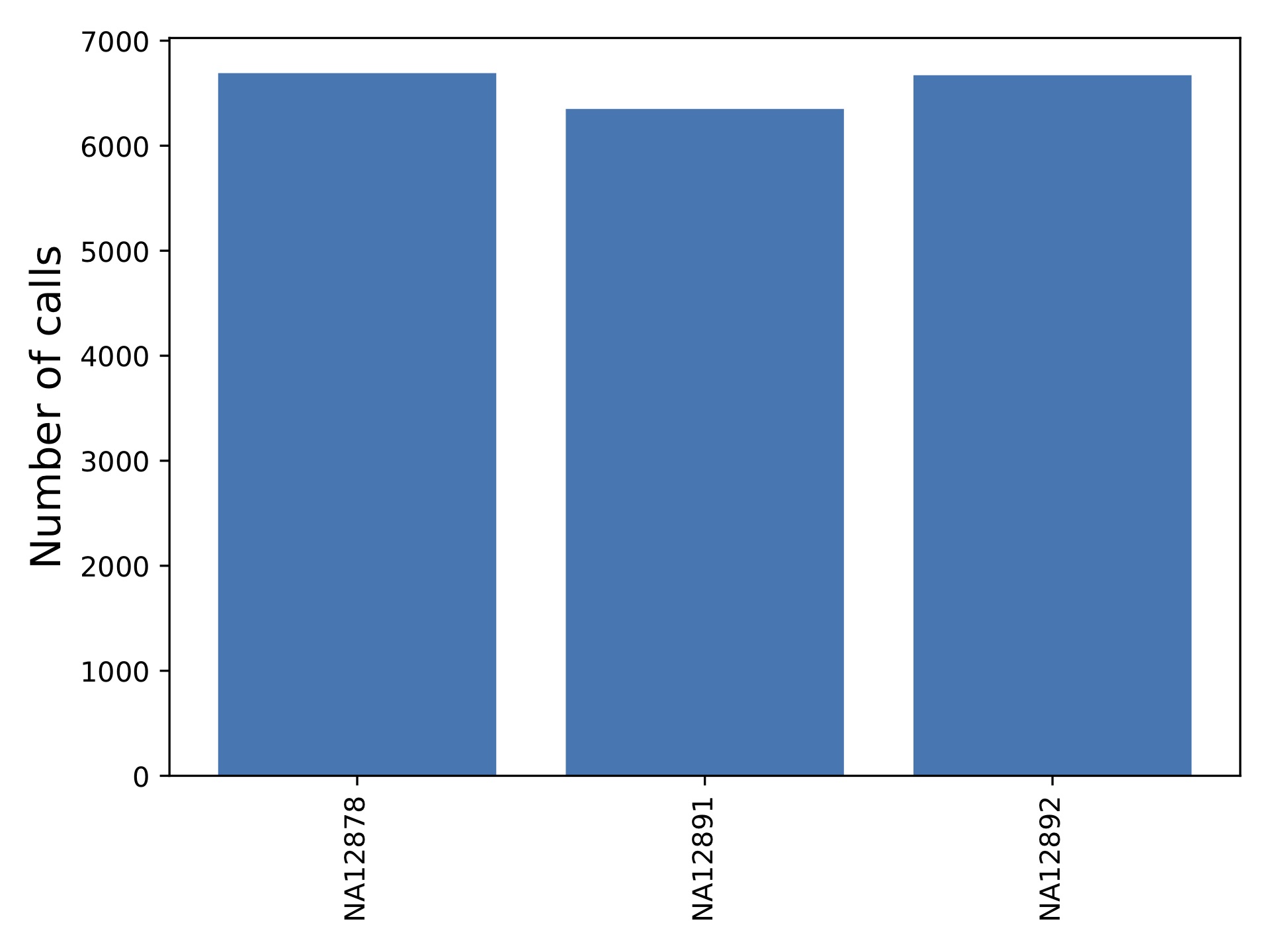
This shows the number of calls per sample.
popstr-qc-diffref-histogram.pdf
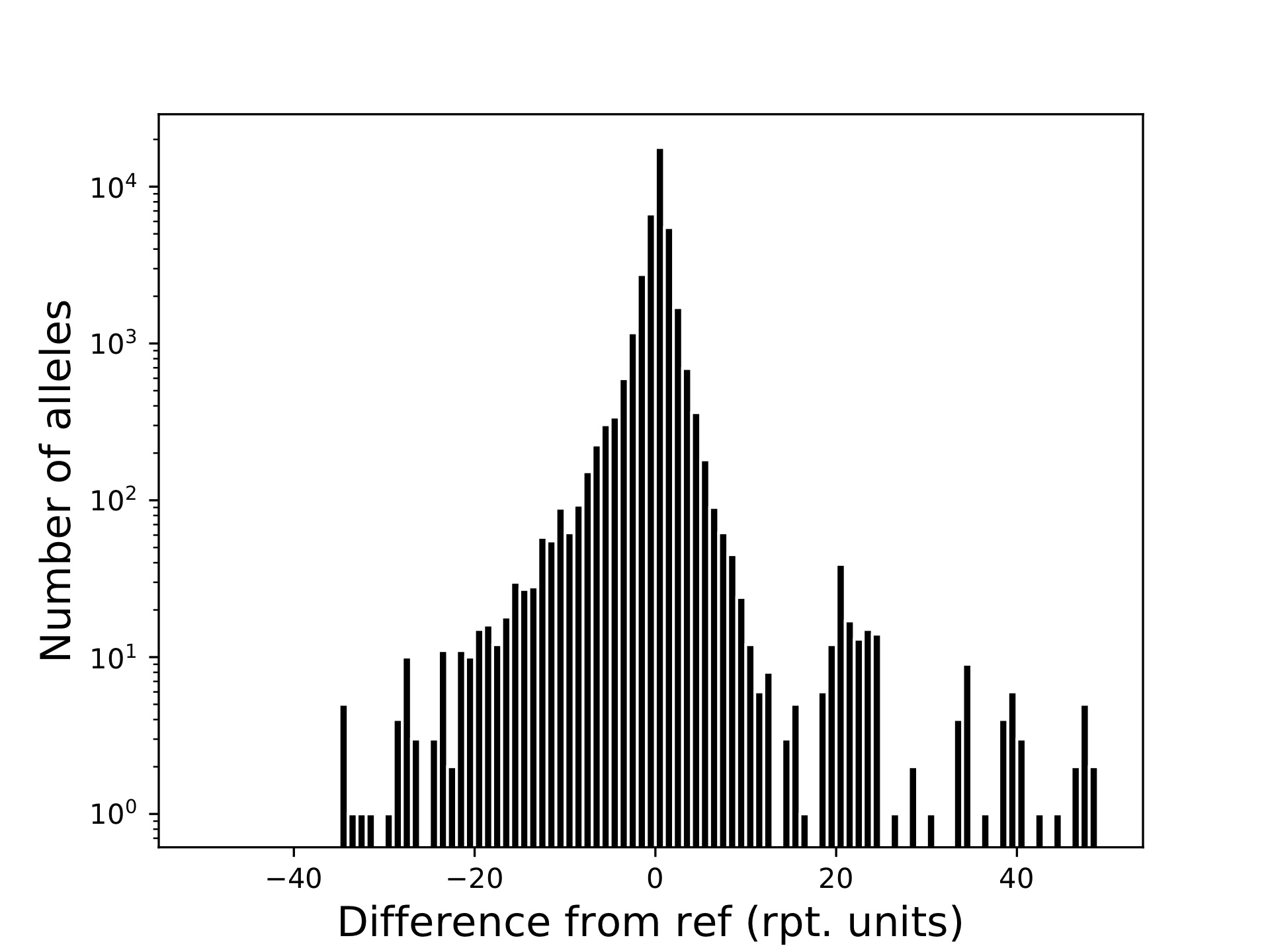
This shows the distribution of allele sizes relative to the reference genome.
popstr-qc-diffref-bias.pdf
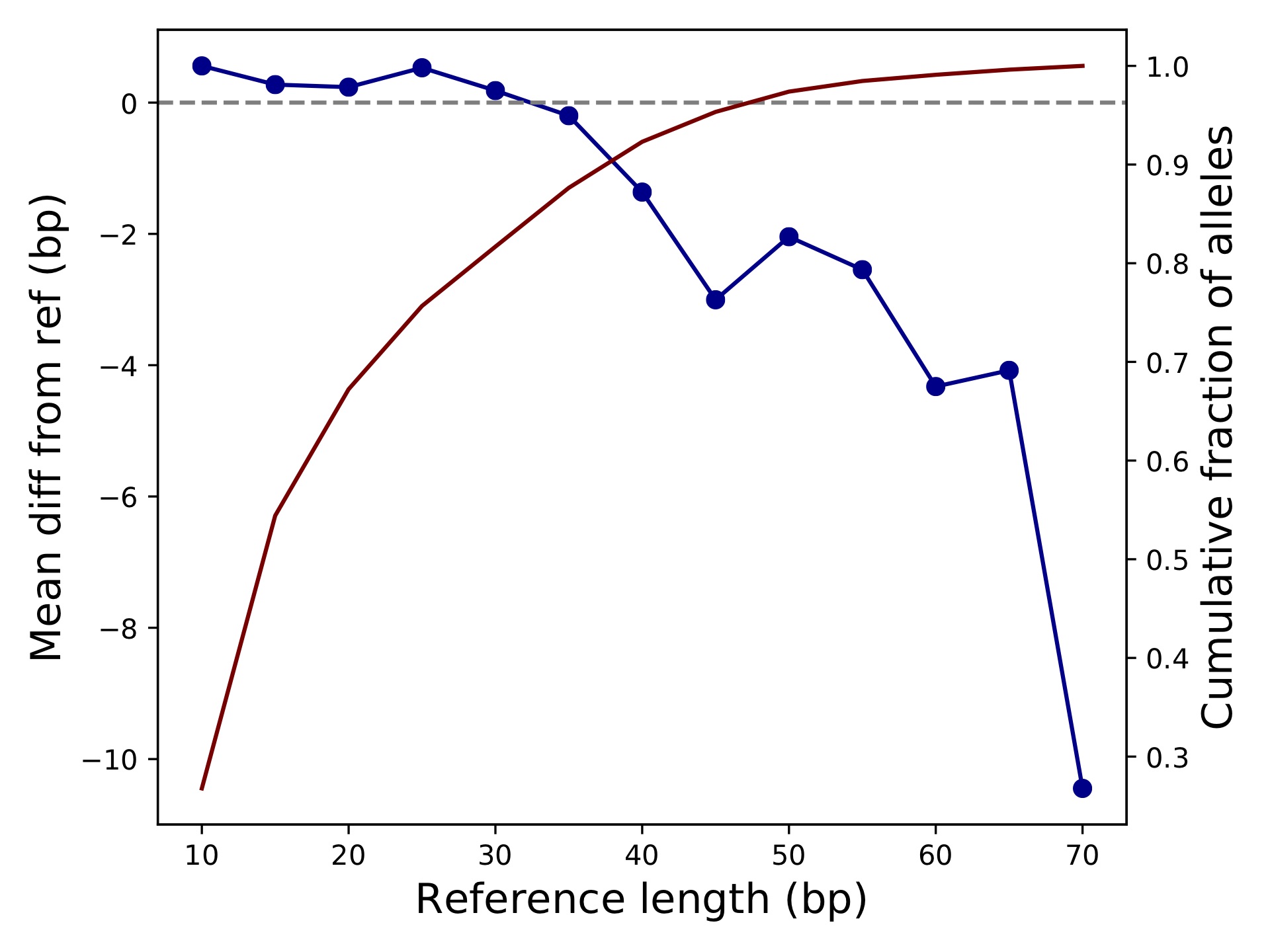
This shows the difference in allele size from the reference as a function of the reference length. We can see as expected calls are biased toward deletions for longer TRs.